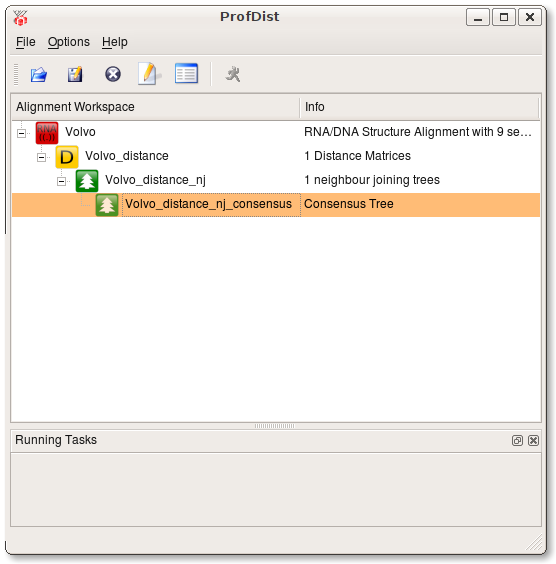
Quickstart
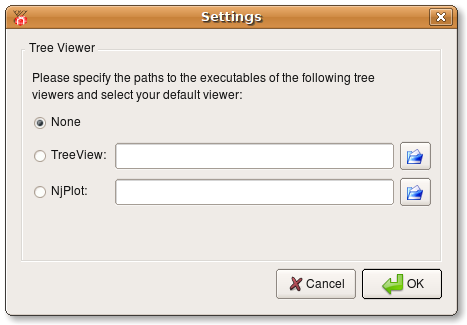
Before you start manipulating alignments make sure you have either TreeView or NJplot installed. These programs are used by ProfDist to view phylogenetic trees. When you start ProfDist go to Options --> Settings and specify the paths to the exetuables of the programs. There is no need to install both viewers. The following screenshot shows the settings dialog where you can specify the paths.
ProfDist provides three ways to transform loaded alignments.
- Profile Neighbour Joining
- Distance --> Neighbour Joining --> Consensus
- Bootstrapping --> Distance calculation --> Neighbour Joining --> Consensus
1. Profile Neighbour Joining
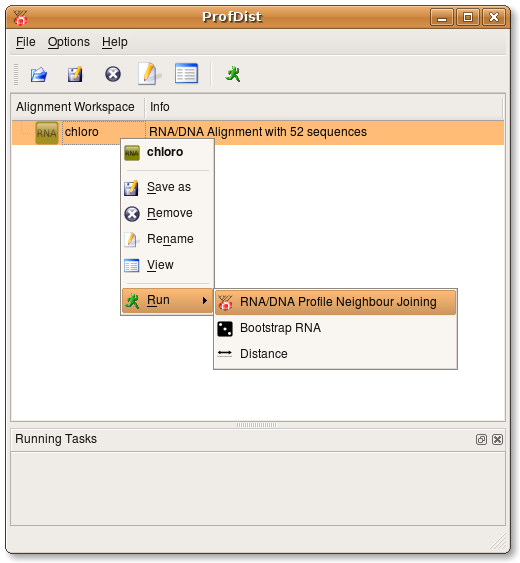
Open an alignment file by pressing the open button ![]() .
After you specified an alignment file you want to open you will be asked what
type of alignment the file contains. You have to choose between the following options:
.
After you specified an alignment file you want to open you will be asked what
type of alignment the file contains. You have to choose between the following options:
- Plain RNA/DNA alignment
- RNA/DNA structure alignment (alignment file with secondary structure information)
- Plain protein alignment
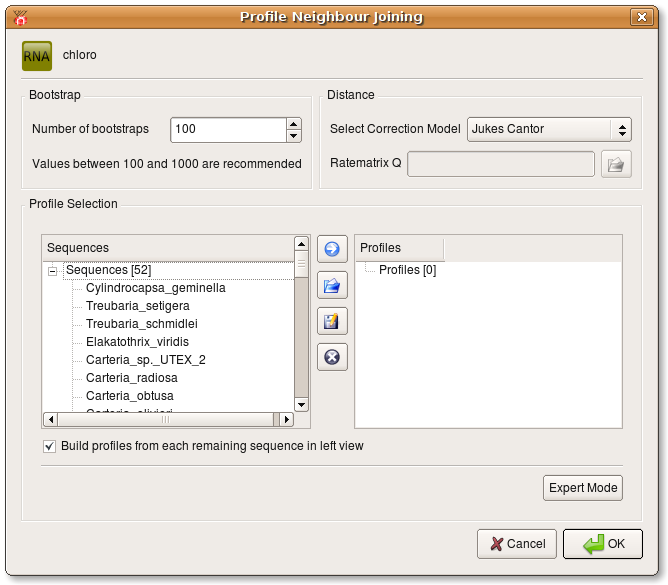
Now a new dialog opens where you can specify the number of bootstraps, the correction model used during distance calculation and you can define profiles from the source alignment which are used during Profile Neighbour Joining. You either have to define at least 3 profiles or select the check box at the bottom of the dialog.
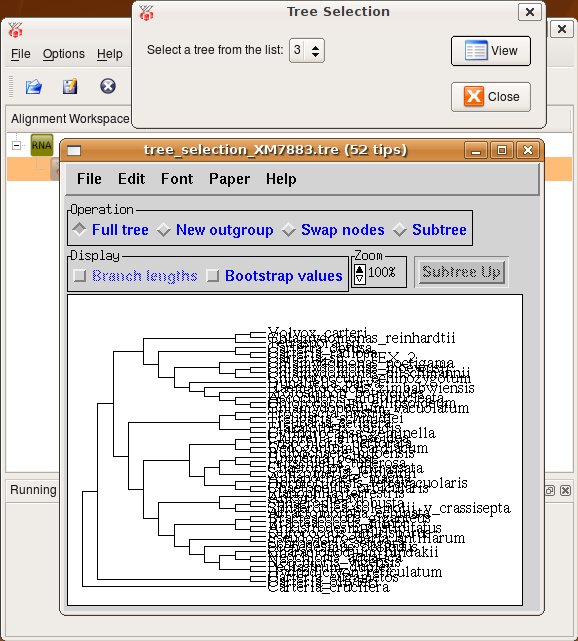
Press Ok and a new task is started. You can track it's progress at the bttom of the ProfDist main window. The resulting data item is then appended as a subitem at the source alignment. If you want to inspect the data item you have to select View from the context menu of this item or select View from the toolbar. A viewer opens and you can see a detailed representation of the data as you can see in the following screenshot.
2. Bootstrapping --> Distance calculation --> Neighbour Joining --> Consensus
This path offers a way to manually perform the operations Bootstrap, Distance Calculation, Neighbour Joining and Consensus on an alignment.
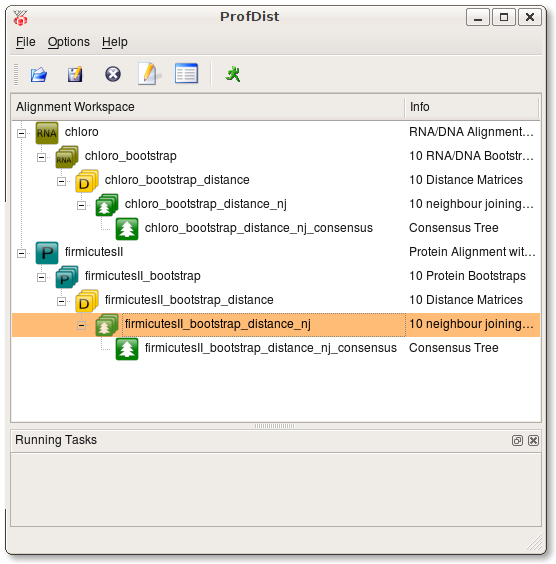
First open an alignment. Then press the right mouse button on this item and navigate to the Run menu and select Bootstrap. After the bootstrap operation has succeeded you will find a new item which is added as a subitem of the source alignment. It is the common behaviour of ProfDist that every resulting data item is added as a subitem to its source item. By this approach a tree of data items is build. This tree very clearly shows which data item the source of other data items is.
3. Distance --> Neighbour Joining --> Consensus
As a third possibility you can directly calculate a distance matrix on an alignment without performing bootstrap on it.